document.querySelector('video').playbackRate = 1.2in bio-informatics or computational-biology
…or a lab in any field of -informatics or computational-
My personal experience felt about like this:

Using a real data-focused IDE!
https://www.spyder-ide.org/
Actually exploring data this real IDE (above),
is in contrast to the later documenting and publishing you do with
different tools (below).
in bio-informatics or computational-biology
…or a lab in any field of -informatics or computational-
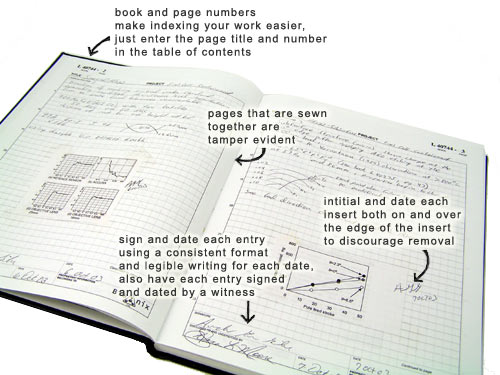
https://en.wikipedia.org/wiki/Lab_notebook
Lab notebooks are a real thing, and scientists actually keep
them!
They are used for documentation, which feeds into publication.
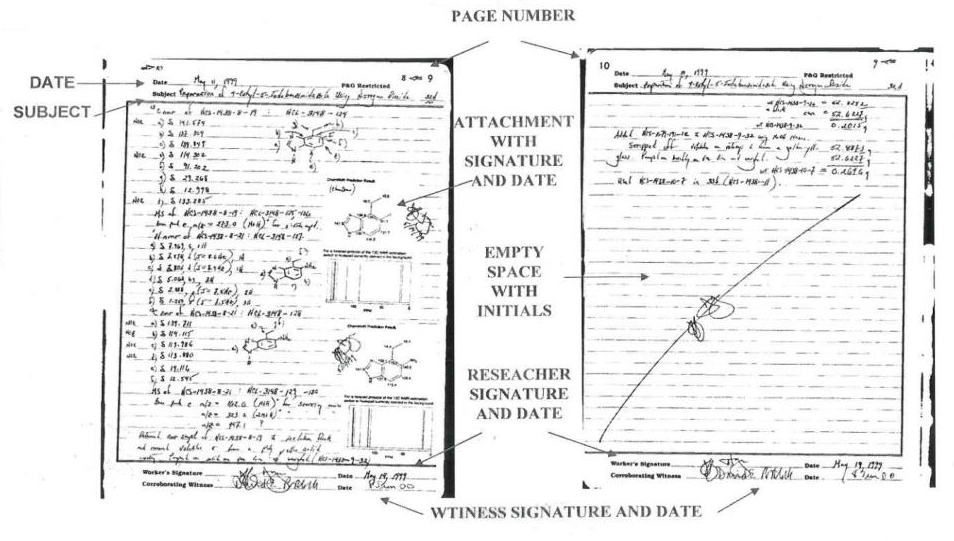
The classic paper model:

How to increase consistency, transparency, and computability in
scientific publishing?
Goal: A communicative, transparent, easy-to-read, and actually executable publication.
A variety of data-focused documentation and publication tools.
https://jupyter.org/
notebooks made this approach popular.
A type of lab-notebook for data analysis, that enabled more easily
reproducible science!
Jupyter notebooks are NOT great for coding, implementation,
early-stage programming,
NOR for real data exploration (in my opinion),
but they were a great start for improving the culture of transparent
publication.
They are nicer for more polished documentation stages of coding data
analysis.
https://towardsdatascience.com/5-reasons-why-you-should-switch-from-jupyter-notebook-to-scripts-cb3535ba9c95
https://www.section.io/engineering-education/why-data-scientists-need-to-move-from-jupyter-notebooks-to-scripts/
https://towardsdatascience.com/5-reasons-why-jupyter-notebooks-suck-4dc201e27086
https://owainkenwayucl.github.io/2017/10/03/WhyIDontLikeNotebooks.html
Despite its widespread popularity, jupyter, and the ipynb
format,
are both buggy and poorly programmed…
Thanks to Jupyter for kickstarting a trend,
and for the great high-level design/concept,
but the back-end implementation was technically bad.
Thus, I no longer recommend using .ipynb as your primary
format
(perhaps a secondary format; see below).
Install
System repos:
There are multiple packages, named differently across distribution, so
search:
$ sudo dnf/apt/zypper search jupyter
For example, install some, or all, of the results of the previous
search:
$ sudo dnf/apt/zypper install jupyter-*
If you want a newer version, don’t have sudo access, or use system
repositories,
then use:
To search:
$ pip3 search jupyter
Choose what you want, and install:
$ pip3 install --upgrade jupyter jupyterlab notebook --user
https://jupytext.readthedocs.io
Provides a nice bandage fix for jupyter’s poor back-end.
Plain-text!
Jupyter notebook convertible!
Many formats, including .py file scripts!
Very picky about which python interpreter it uses, on Linux for example,
with system python exists.
Install with:
https://jupytext.readthedocs.io/en/latest/install.html
#!/bin/bash
pip3 install --upgrade --force jupyter jupyterlab notebook jupytext --user
# To see the menu in jupyterlab:
jupyter labextension enable jupyterlab-jupytext
# Or, on older versions of notebook:
jupyter serverextension enable jupytext --user
jupyter nbextension install --py jupytext --user
jupyter nbextension enable --py jupytext --userhttps://quarto.org
Quarto intends to do things right, from the start, instead of creating a
bandage.
Plain-text.
Jupyter notebook convertible!
Independent format, rather than just tailing jupyter, as Jupytext
does.
Uses the the amazing pandoc (below).
A bit too R-friendly/focused… visual editor only in RStudio (a nice IDE
for a lousy language, R).
https://jupyterbook.org
Jupyter-book builds on the previous two, intended for for larger
publications.
Similar to jupytext, but with more book-like features, a layer “above”
jupytext.
https://pandoc.org/
Pandoc markdown preceded all of these, and was light-years ahead of its
time!
More general purpose than above, for converting between MANY MANY
formats, including jupyter notebooks.
Back-end for some of the projects above.
Amazing project, clean code, awesome!!
https://www.sagemath.org/
is a better alternative to the proprietary Wolfram-Mathematica.
Not bio/data-specific, but just another scientific/mathematical
programming notebook system.
See: ../../DataStructuresLab/Content/03-VersionControl.html
Use a version-controlled repository to archive versions of your
progress, and to publish data, code, and results!
Often, you want the entire experiment to be reproducible, by another
lab or person!
Hiding your own data or code is counter to the scientific process!
See: ../../DataStructuresLab/Content/00-VirtualMachines.html
As a computer scientist into security, I am very much an enthusastic
fan of Virtual Machines in general, though our book for biologists who
want to do computation better ( https://www.biostarhandbook.com/ ) also suggests VMs as
a way to bundle up an environment for replicability:
1. Do a real science study,
2. write the scripts that go from raw data to final figures (with no
human input!!),
3. put the data and scripts in a VM,
4. run them all,
5. generate the figures and paper, etc.
6. Then right when you finish, export the VM as an OVA/snapshot (e.g.,
https://www.virtualbox.org/), and publish it all.
Afterwards, there is no question about what produced the results, and
anyone can do so.
This is both good proactive future-proof science,
but also good defensive strategy if your results are being replicated by
another researcher.
Publishing a git repository of your work has become the gold standard
of public code/data.
Sites like https://mybinder.org/ enables you to run the jupyter
notebooks in an arbitrary Git repository on someone-else’s computer,
while installing a pre-specified dependency environment.
One problem is that you can’t securely perform any push operations back
to that Git repository you are working on, without disclosing your
password to the owner of the binder server…
Future-proof dependency handling is much less reliable than a VM.
Ask:
Is this as future-proof and standalone reproducible as a VM?
Is it as easy as a VM?
Is it as likely to be reproduced?