document.querySelector('video').playbackRate = 1.2“Wherever there is an adaptation that is highly successful in
a broad range of similar environments,
it is apt to emerge again and again, independently -
the phenomenon known in biology as convergent evolution.
I call these adaptations ‘good tricks.’“
- Daniel Dennett (A thought-leader in evolution, and a great writer)
A perspective-changing read by Dennet and Levin:
https://aeon.co/essays/how-to-understand-cells-tissues-and-organisms-as-agents-with-agendas

The development of protein-sequencing methods start with Sanger and
Tuppy in 1951.
This led to the sequencing of representatives of several of the more
common protein families,
such as cytochromes, from a variety of organisms.
Margaret Dayhoff (1972, 1978) and collaborators at the National
Biomedical Research Foundation (NBRF), Washington, DC,
were the first to assemble databases of these sequences into a protein
sequence atlas in the 1960s,
and their collection center eventually became known as the Protein
Information/Identification Resource (PIR).
Dayhoff and her coworkers organized the proteins into families and
super-families,
based on the degree of sequence similarity.
This was more objective than inferring evolutionary relationships based
on physiological form and structure.
For example:
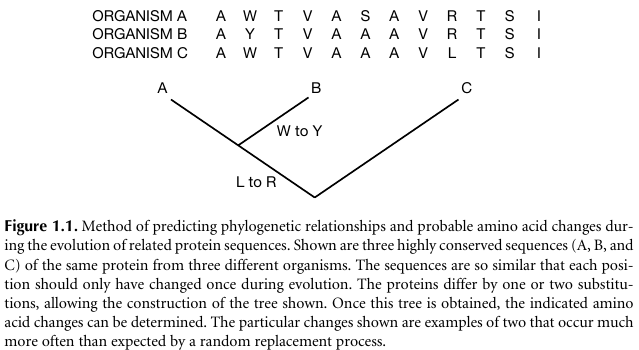
How many differences between:
A and B?
A and C?
B and C?
How to construct such a tree structure?
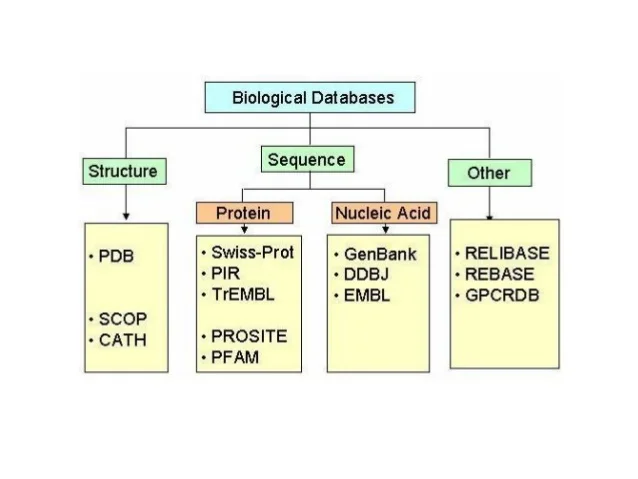
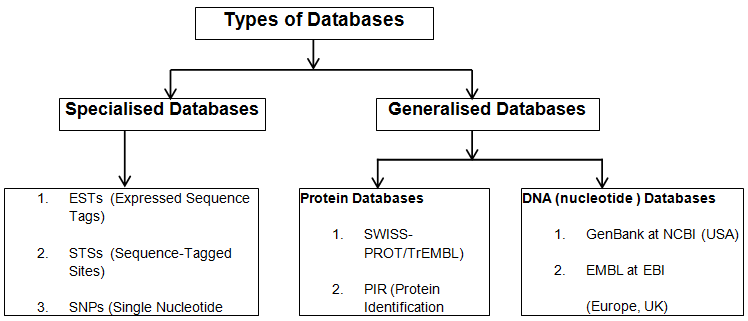
DNA sequence databases were first assembled at:
the Los Alamos National Laboratory (LANL), New Mexico, by Walter Goad
and colleagues in the GenBank database, and
the European Molecular Biology Laboratory (EMBL) in Heidelberg,
Germany.
Initially, a sequence entry included a computer filename, and DNA or
protein sequence files.
These were eventually expanded to include much more information about
the sequence,
such as function, mutations, encoded proteins, regulatory sites, and
references.
This information was then placed, along with the sequence, into a
database format,
that could be readily searched for many types of information.
 ]
]
An important step in providing sequence database access was the
development of Web pages,
that allow queries to be made of the major sequence databases (GenBank,
EMBL, etc.).
An early example of this technology at NCBI was a menu-driven program
called GEN-INFO,
developed by D. Benson, D. Lipman, and colleagues.
This program searched rapidly through previously indexed sequence
databases,
for entries that matched a biologist’s query.
Subsequently, NCBI created a derivative program called ENTREZ,
with a simple window-based interface, and eventually a Web-based
interface.
The idea behind these programs was to provide an easy-to-use
interface,
with a flexible search procedure to the sequence databases.
Because DNA sequencing involves ordering a set of peaks (A, G, C, or T)
on a sequencing gel,
the process can be quite error-prone, depending on the quality of the
data.
As more DNA sequences became available in the late 1970s,
interest also increased in developing computer programs to analyze these
sequences.
In 1982 and 1984, Nucleic Acids Research published two special
issues,
devoted to the application of computers for sequence analysis,
including programs for large mainframe computers down to the then-new
microcomputers.

In 1970, A.J. Gibbs and G.A. McIntyre (1970) described a new
method.
The method compared two amino acid and nucleotide sequences, in which a
graph was drawn,
with one sequence written across the page, and the other down the
left-hand side.
Whenever the same letter appeared in both sequences,
a dot was placed at the intersection of the corresponding sequence
positions on the graph
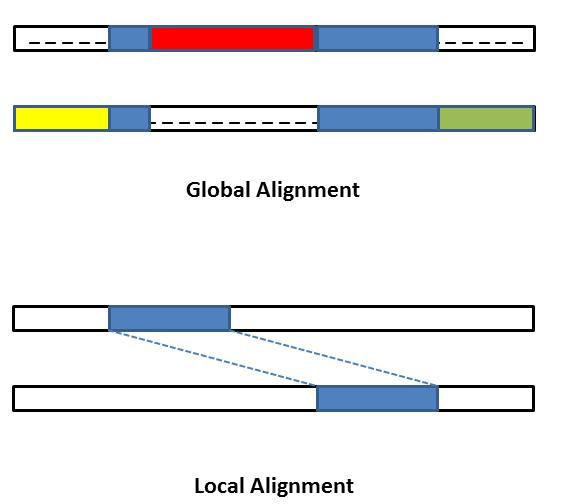
There are various methods for aligning:
entire matching segments,
small matching adjacent segments, and
multiple variable-length segments.
Global versus local alignment:
Also, multiple sequence alignment:
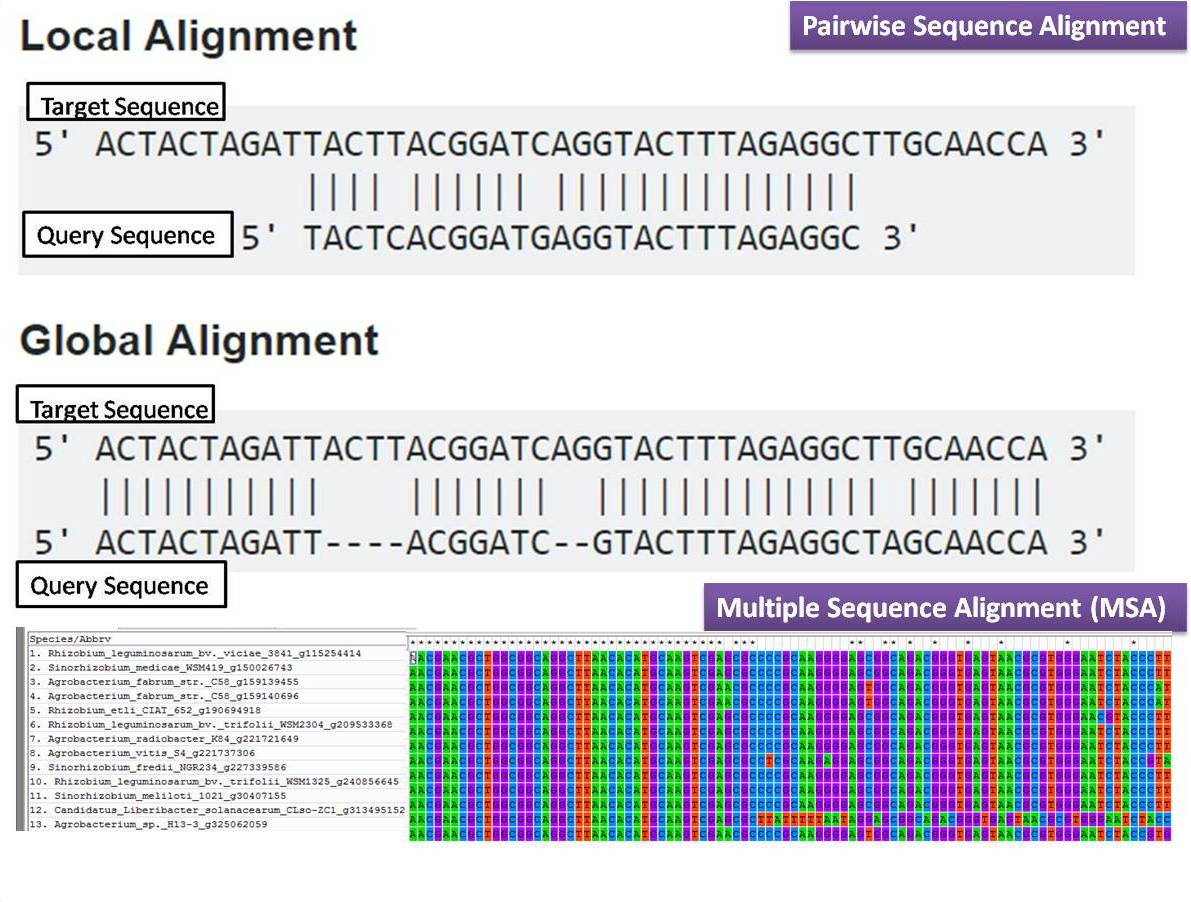
Why is this useful?
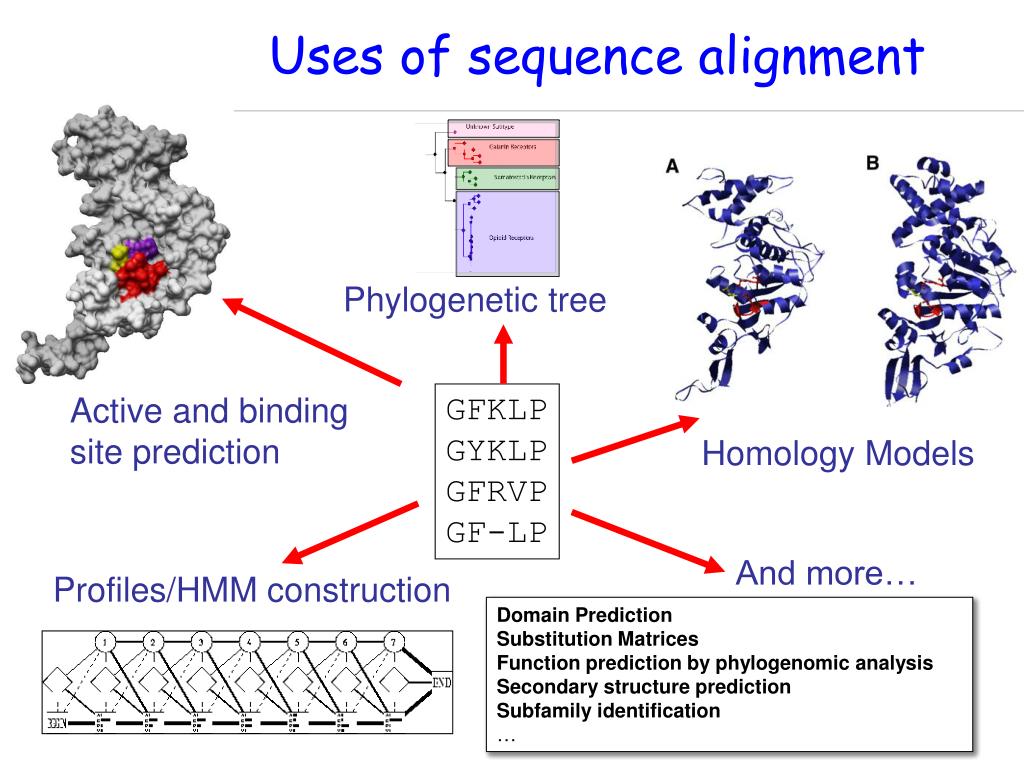
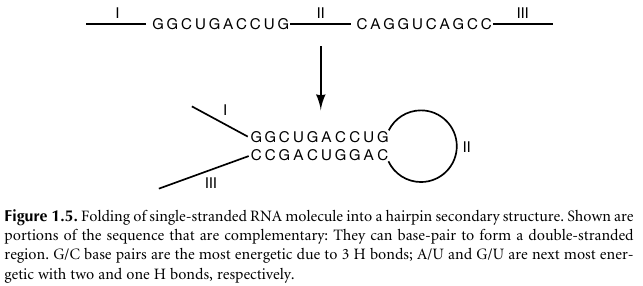
Methods for predicting RNA secondary structure on computers were also
developed at an early time.
For example, if the complement of a sequence on an RNA molecule is
repeated,
further down the sequence, in the opposite chemical direction,
then the regions may base-pair and form a hairpin structure:
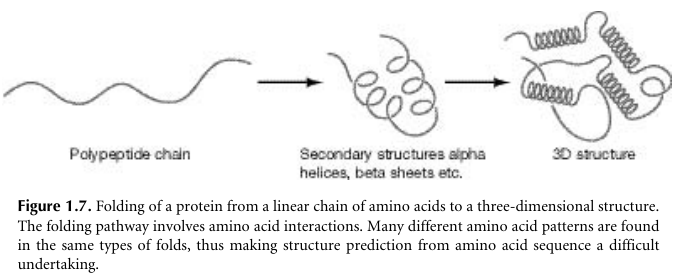
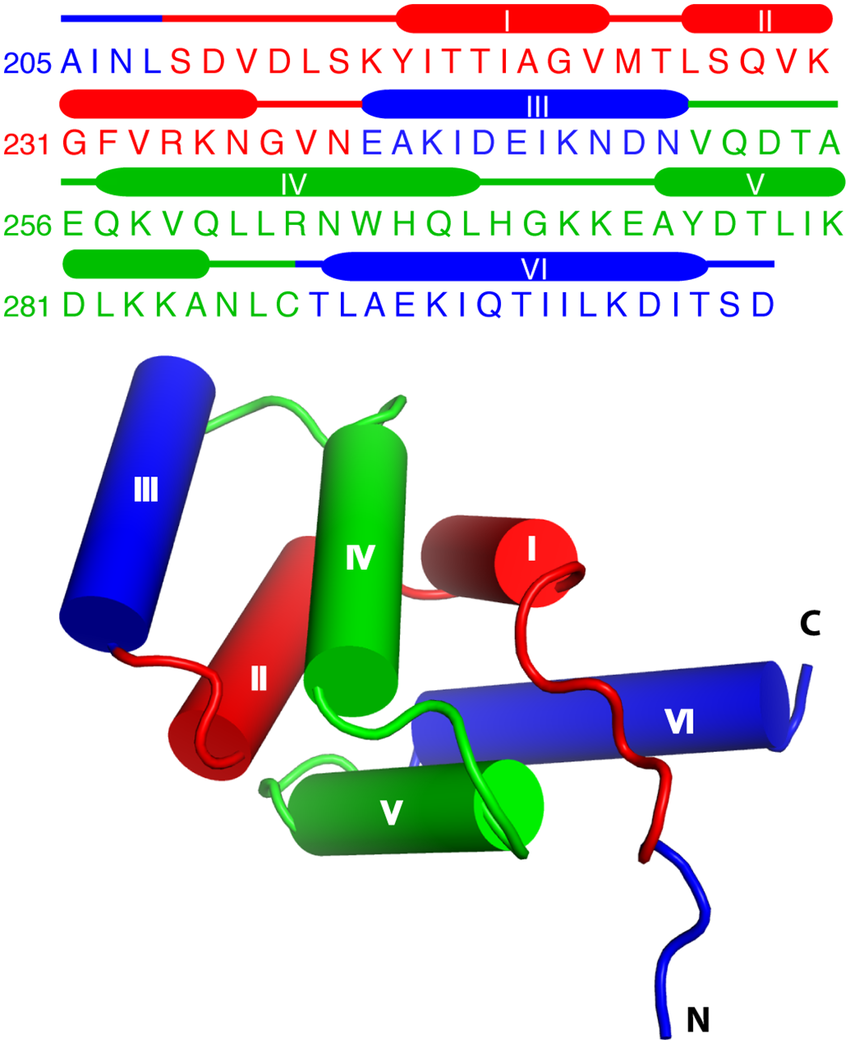
There are a large number of proteins whose sequences are known,
but very few whose structures have been solved.
Solving protein structures involves the time-consuming and highly
specialized procedures,
of X-ray crystallography and nuclear magnetic resonance (NMR).
There we try to predict the structure of a protein, given its
sequence.
Early attempts were made at predicting protein structure from
sequence.
+++++++++++++++++++ Cahoot-05-1
Variations within a family of related nucleic acid or protein
sequences,
provide a source of information for evolutionary biology,
enabling the discovery of relationships between species in an
objectively quantifiable manner.
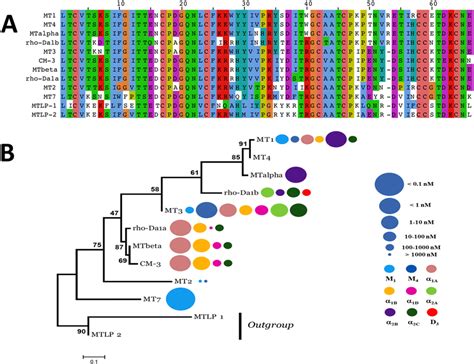
It’s not just species that one can compare,
but also proteins within an organism,
which can be duplicated within an organism,
and then re-purposed for new, independent functions.
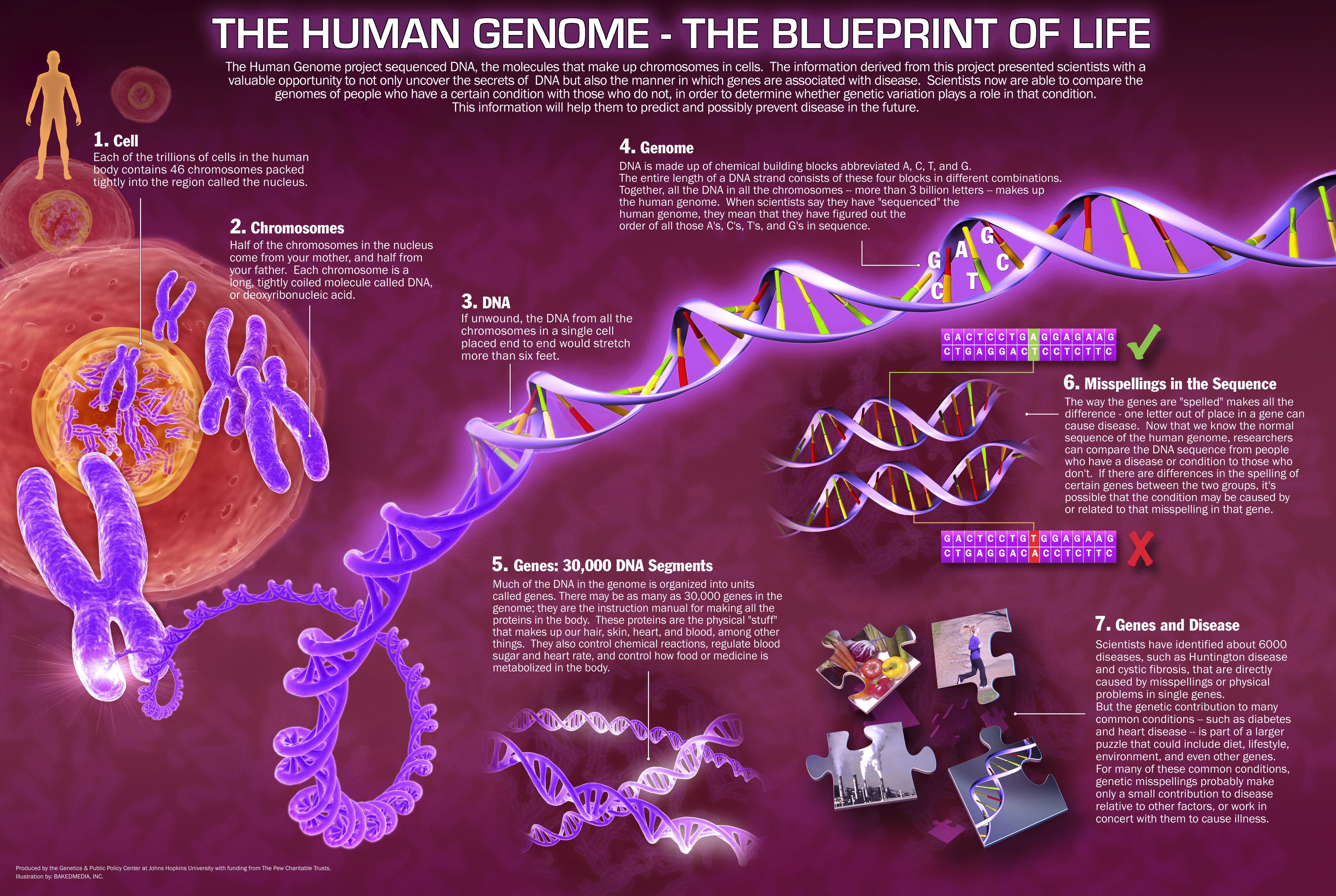
The first genome database, was called ACEDB (a C. elegans
database),
The methods to access this database were developed by (Cherry and
Cartinhour 1993).
This database was accessible through the internet and allowed retrieval
of sequences,
information about genes and mutants, investigator addresses, and
references.
Similar databases were subsequently developed for A. thaliana and S.
cerevisiae.
This is C. elegans:
And then the field of bioinformatics exploded
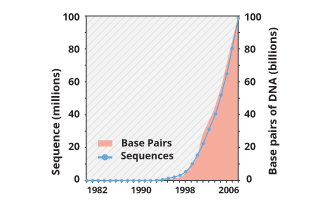
“… from 1982 to the present, the number of bases in GenBank has doubled
approximately every 18 months”. As of 15 August 2017, GenBank release
221.0 has:
203,180,606 loci,
240,343,378,258 bases,
from 203,180,606 reported sequences.
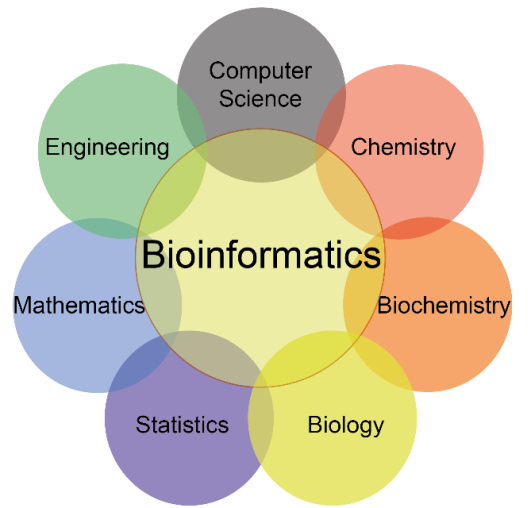
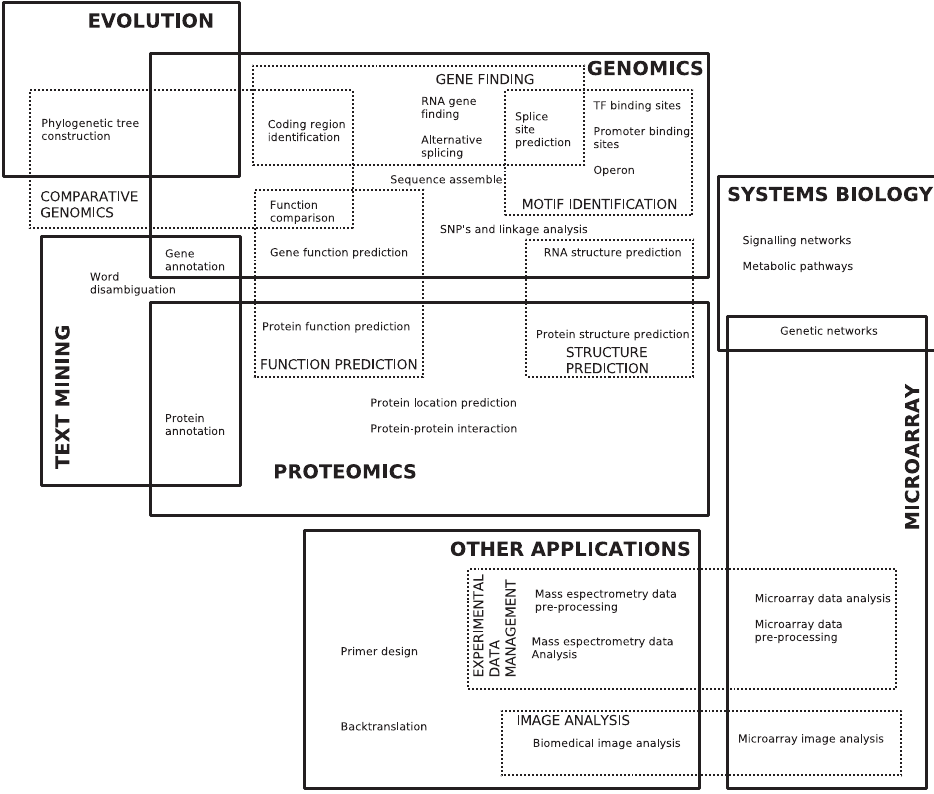
Venn of the nexus of many fields
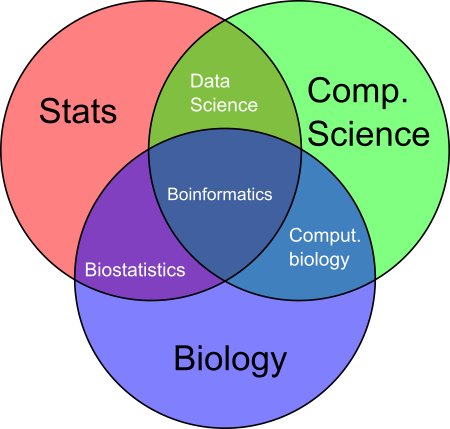
Contrasted to data science
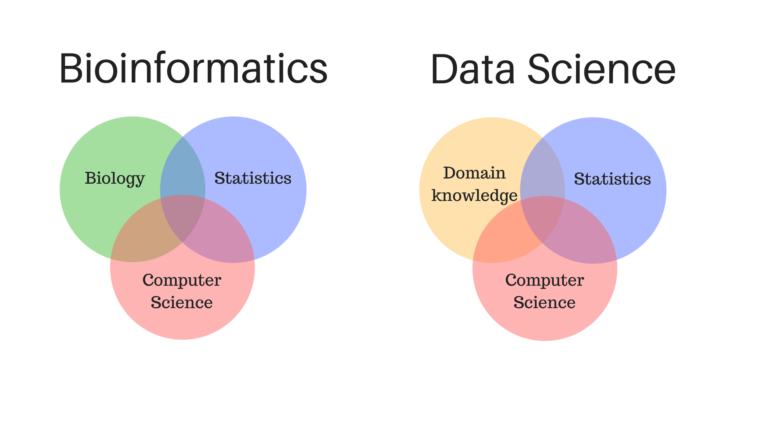
Same job, way worse pay…
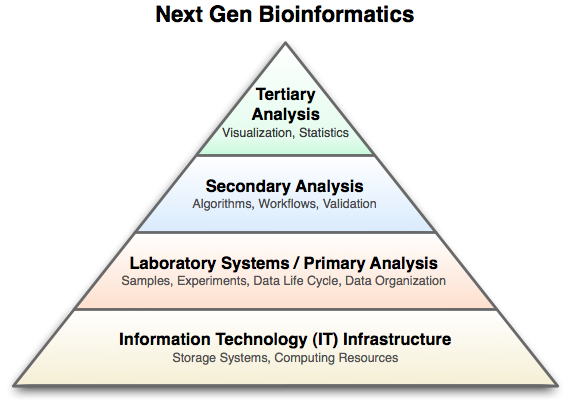
Slightly more detail
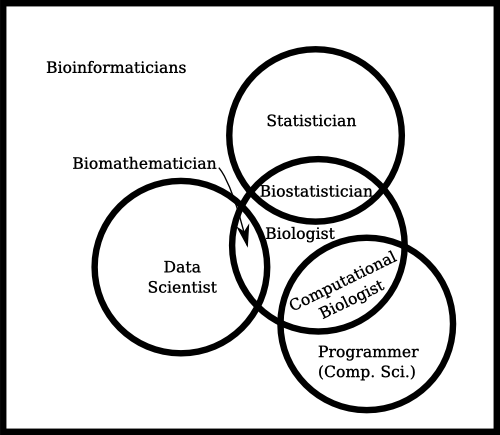
Even more detail
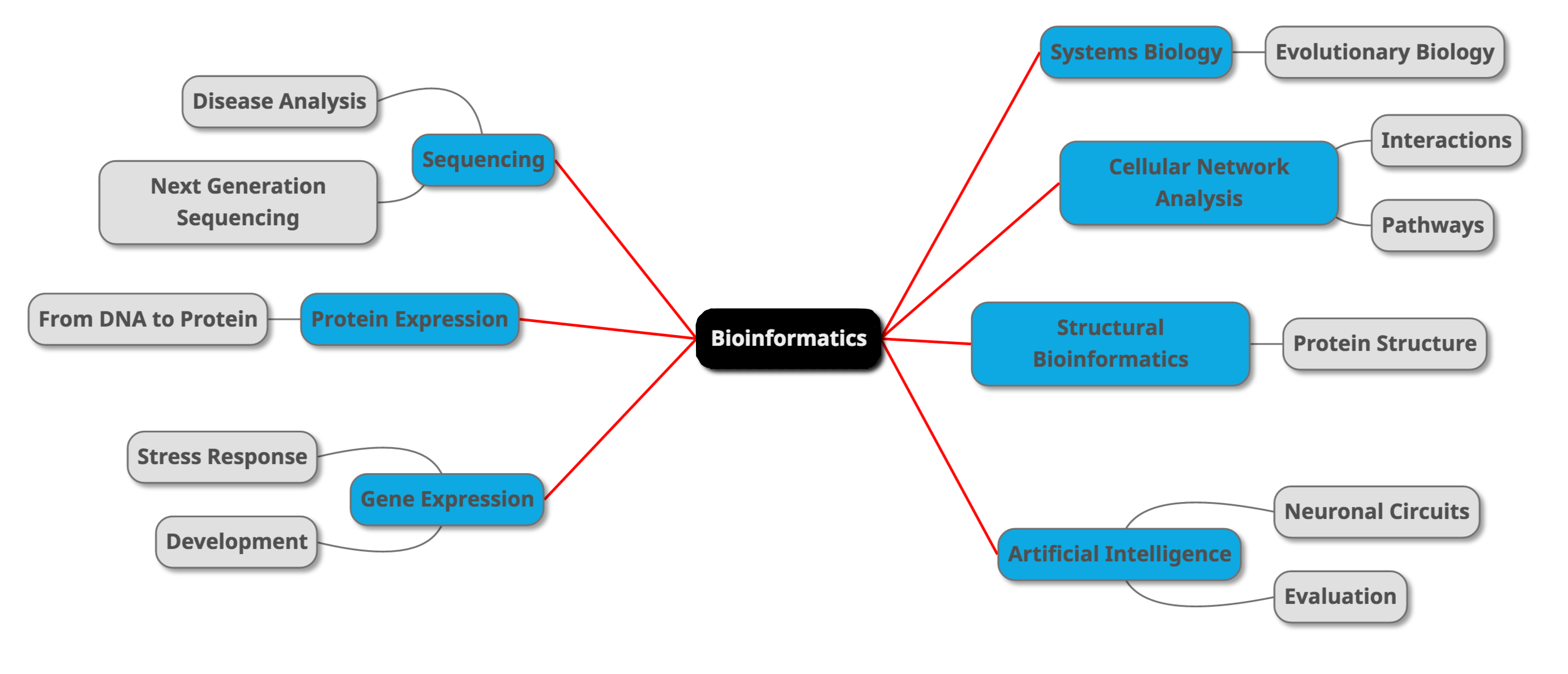
A different perspective
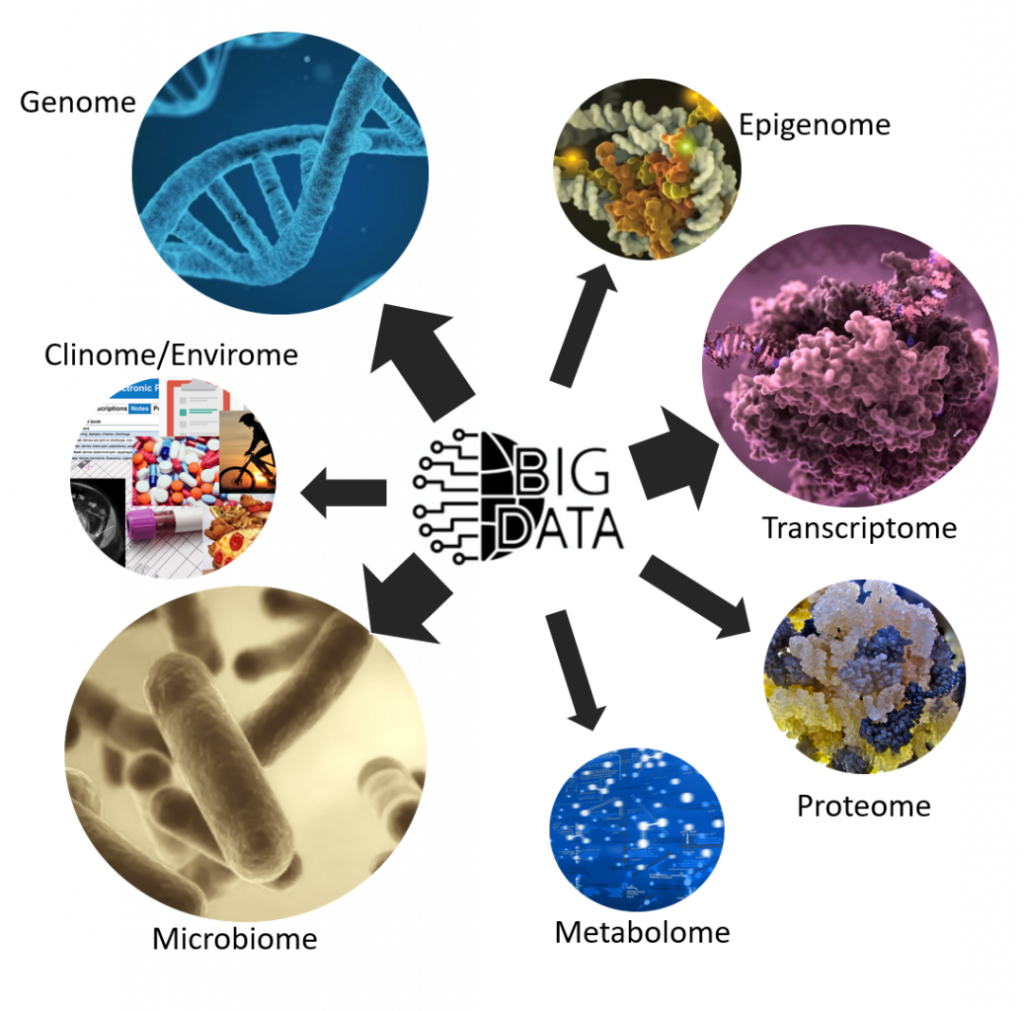
AI methods in Bioinformatics
https://en.wikipedia.org/wiki/Computational_epidemiology
https://en.wikipedia.org/wiki/Mathematical_modelling_of_infectious_disease
https://en.wikipedia.org/wiki/Compartmental_models_in_epidemiology
https://en.wikipedia.org/wiki/Computational_biology
https://en.wikipedia.org/wiki/Bioinformatics
https://en.wikipedia.org/wiki/Sequence_assembly
https://en.wikipedia.org/wiki/Sequence_analysis
https://en.wikipedia.org/wiki/Comparative_genomics
https://en.wikipedia.org/wiki/Health_informatics
https://en.wikipedia.org/wiki/Imaging_informatics
https://en.wikipedia.org/wiki/Neuroinformatics
https://en.wikipedia.org/wiki/Computational_neuroscience
https://en.wikipedia.org/wiki/Modelling_biological_systems
https://en.wikipedia.org/wiki/Computational_phylogenetics
https://en.wikipedia.org/wiki/Computational_genomics
https://en.wikipedia.org/wiki/Biodiversity_informatics
https://en.wikipedia.org/wiki/Biological_network
https://en.wikipedia.org/wiki/Structural_bioinformatics
https://en.wikipedia.org/wiki/Ecosystem_model
https://en.wikipedia.org/wiki/Models_of_DNA_evolution
https://en.wikipedia.org/wiki/Translational_bioinformatics
https://en.wikipedia.org/wiki/Gene_ontology
https://en.wikipedia.org/wiki/Gene_prediction
https://en.wikipedia.org/wiki/Bioimage_informatics
https://en.wikipedia.org/wiki/Protein_structure_prediction
https://en.wikipedia.org/wiki/Computational_anatomy
https://en.wikipedia.org/wiki/Cellular_model
https://en.wikipedia.org/wiki/Computational_biology
https://en.wikipedia.org/wiki/Bioinformatics
https://en.wikipedia.org/wiki/Sequence_assembly
https://en.wikipedia.org/wiki/Sequence_analysis
https://en.wikipedia.org/wiki/Comparative_genomics
https://en.wikipedia.org/wiki/Health_informatics
https://en.wikipedia.org/wiki/Imaging_informatics
https://en.wikipedia.org/wiki/Neuroinformatics
https://en.wikipedia.org/wiki/Computational_neuroscience
https://en.wikipedia.org/wiki/Modelling_biological_systems
https://en.wikipedia.org/wiki/Computational_phylogenetics
https://en.wikipedia.org/wiki/Computational_genomics
https://en.wikipedia.org/wiki/Biodiversity_informatics
https://en.wikipedia.org/wiki/Structural_bioinformatics
https://en.wikipedia.org/wiki/Ecosystem_model
https://en.wikipedia.org/wiki/Models_of_DNA_evolution
https://en.wikipedia.org/wiki/Translational_bioinformatics
https://en.wikipedia.org/wiki/Gene_ontology
https://en.wikipedia.org/wiki/Gene_prediction
https://en.wikipedia.org/wiki/Bioimage_informatics
https://en.wikipedia.org/wiki/Protein_structure_prediction
https://en.wikipedia.org/wiki/Computational_anatomy
https://en.wikipedia.org/wiki/Cellular_model
+++++++++++++++++++
Discussion question:
Databases help us compute on raw data.
Ontologies help us compute on verbal knowledge, and meta-data, and
interpretation.
They are the basis of many modern language models.
What might you envision that cutting edge language models might
contribute to to the scientific literature in bioinformatics?
Benefits? Costs?
https://www.mkbergman.com/374/an-intrepid-guide-to-ontologies/
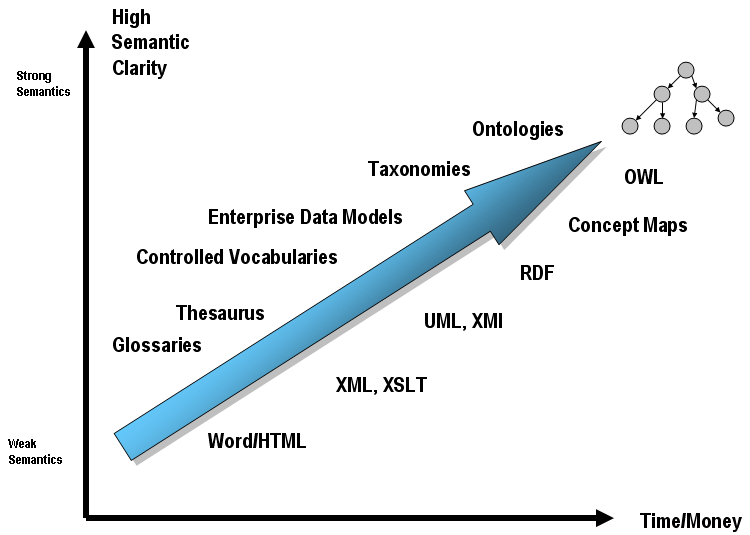
An ontology is a formal naming and definition of:
the types, properties, and interrelationships of the entities,
that really exist in a particular domain of discourse.
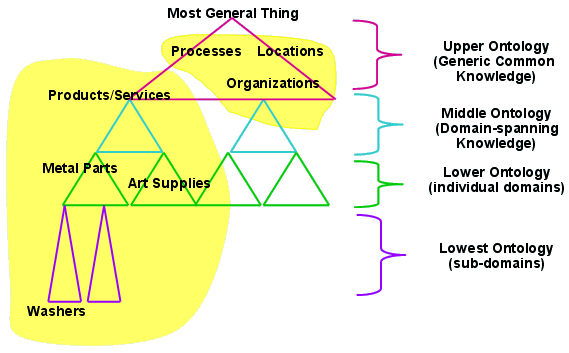
An upper ontology (or foundation ontology) is a model
of the common objects,
that are generally applicable across a wide range of domain
ontologies.
It usually employs a core glossary that contains terms and associated
object descriptions,
as they are used in various relevant domain sets, for example, the Basic
Formal Ontology (BFO)
Domain ontology example:
Open Biomedical Ontologies (abbreviated OBO; formerly Open Biological
Ontologies).
Create controlled vocabularies for shared use across different
biological and medical domains.
As of 2006, OBO forms part of the resources of the U.S. National Center
for Biomedical Ontology,
where it will form a central element of the NCBO’s BioPortal.
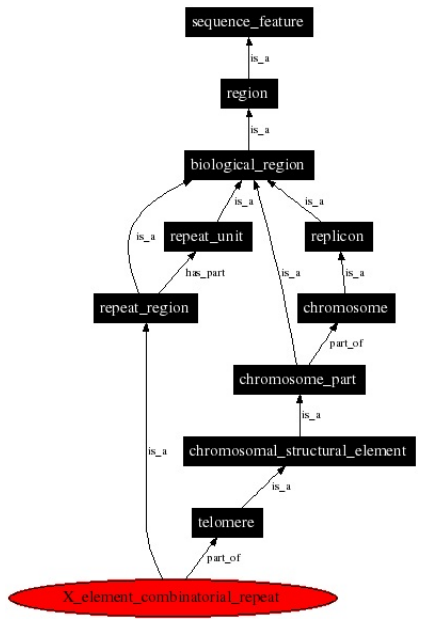
The Sequence Ontology (SO)
http://www.sequenceontology.org
SO defines sequence features used in biological sequence
annotation.
For example:
An X element combinatorial repeat, is a repeat region,
located between the X element, and the telomere or adjacent Y’
element.
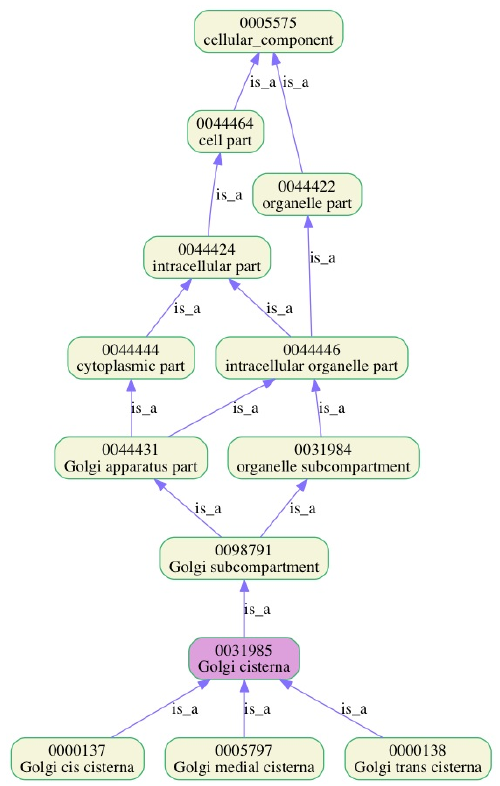
The Gene Ontology (GO) is a controlled vocabulary that connects each
gene to one or more functions.
http://geneontology.org/
GO is intended to categorize gene products, rather than the genes
themselves.
Different products of the same gene may play very different roles,
and labelling and treating all of these functions under the same gene
name
may (and often does) lead to confusion.
++++++++++++ Cahoot-05-2
https://en.wikipedia.org/wiki/List_of_biological_databases
https://en.wikipedia.org/wiki/List_of_biodiversity_databases
https://en.wikipedia.org/wiki/List_of_neuroscience_databases
Many ore to come later!